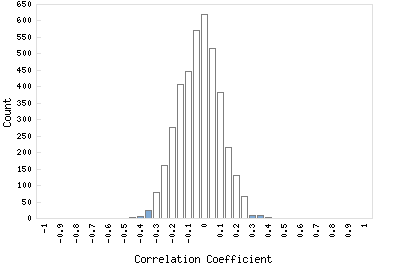
Correlation Catalog M. tuberculosis H37Rv: Rv3752c - cytidine/deoxycytidylate deaminase
Distribution of gene correlation: Rv3752c vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0973c | acetyl-/propionyl-CoA carboxylase alpha subunit accA2 |

|
 -0.41099 -0.41099
| |
| Rv3093c | oxidoreductase |

|
 -0.40471 -0.40471
| |
| Rv2417c | conserved hypothetical protein |

|
 -0.38693 -0.38693
| |
| Rv0085 | hydrogenase hycP |

|
 -0.37556 -0.37556
| |
| Rv1293 | diaminopimelate decarboxylase lysA |

|
 -0.37228 -0.37228
| |
| Rv1295 | threonine synthase thrC |

|
 -0.36217 -0.36217
| |
| Rv3003c | acetolactate synthase large subunit ilvB1 |
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/E.png)
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/H.png)
|
 -0.35725 -0.35725
| |
| Rv1127c | pyruvate, phosphate dikinase ppdK |

|
 -0.35327 -0.35327
| |
| Rv0860 | fatty-acid oxidation protein fadB |

|
 -0.35074 -0.35074
| |
| Rv1595 | L-aspartate oxidase nadB |

|
 -0.33892 -0.33892
| |
| Rv1381 | dihydroorotase pyrC |

|
 -0.33705 -0.33705
| |
| Rv0463 | conserved membrane protein |

|
 -0.33063 -0.33063
| |
| Rv1431 | conserved membrane protein |

|
 -0.32581 -0.32581
| |
| Rv1301 | conserved hypothetical protein |

|
 -0.32346 -0.32346
| |
| Rv1797 | conserved hypothetical protein |

|
 -0.32121 -0.32121
| |
| Rv1583c | phiRv1 phage protein |

|
 -0.31809 -0.31809
| |
| Rv3535c | acetaldehyde dehydrogenase |

|
 -0.31459 -0.31459
| |
| Rv1315 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase murA |

|
 -0.31355 -0.31355
| |
| Rv2933 | phenolpthiocerol synthesis type-I polyketide synthase ppsC |

|
 -0.31345 -0.31345
| |
| Rv0513 | conserved membrane protein |

|
 -0.31195 -0.31195
| |
| Rv3001c | ketol-acid reductoisomerase ilvC |


|
 -0.30973 -0.30973
| |
| Rv3653 | PE-PGRS family protein |

|
 -0.30911 -0.30911
| |
| Rv1837c | malate synthase G glcB |

|
 -0.30863 -0.30863
| |
| Rv1209 | conserved hypothetical protein |

|
 -0.30846 -0.30846
| |
| Rv2509 | short-chain type dehydrogenase/reductase |

|
 -0.30818 -0.30818
| |
| Rv2345 | conserved membrane protein |

|
 -0.30785 -0.30785
| |
| Rv3017c | Esat-6 like protein esxQ |

|
 -0.30766 -0.30766
| |
| Rv0525 | conserved hypothetical protein |

|
 -0.3058 -0.3058
| |
| Rv2134c | conserved hypothetical protein |

|
 -0.30393 -0.30393
| |
| Rv3167c | transcriptional regulator, tetR-family |

|
 -0.30107 -0.30107
| |
| Rv1178 | aminotransferase |

|
 -0.30091 -0.30091
| |
| Rv3019c | Esat-6 like protein esxR |

|
 -0.30001 -0.30001
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3750c | excisionase |

|
 0.43527 0.43527
| |
| Rv2632c | conserved hypothetical protein |

|
 0.43225 0.43225
| |
| Rv2026c | conserved hypothetical protein |

|
 0.39011 0.39011
| |
| Rv1444c | hypothetical protein |

|
 0.37132 0.37132
| |
| Rv2020c | conserved hypothetical protein |

|
 0.36979 0.36979
| |
| Rv3642c | hypothetical protein |

|
 0.36209 0.36209
| |
| Rv3390 | lipoprotein lpqD |

|
 0.35811 0.35811
| |
| Rv1892 | membrane protein |

|
 0.35491 0.35491
| |
| Rv0108c | hypothetical protein |

|
 0.3546 0.3546
| |
| Rv0748 | conserved hypothetical protein |

|
 0.35129 0.35129
| |
| Rv1918c | PPE family protein |

|
 0.33362 0.33362
| |
| Rv2840c | conserved hypothetical protein |

|
 0.32883 0.32883
| |
| Rv0680c | conserved membrane protein |

|
 0.32847 0.32847
| |
| Rv3822 | conserved hypothetical protein |

|
 0.32757 0.32757
| |
| Rv1906c | conserved hypothetical protein |

|
 0.32435 0.32435
| |
| Rv1882c | short-chain type dehydrogenase/reductase |


|
 0.31399 0.31399
| |
| Rv3172c | hypothetical protein |

|
 0.31099 0.31099
| |
| Rv0322 | UDP-glucose 6-dehydrogenase udgA |

|
 0.30869 0.30869
| |
| Rv2302 | conserved hypothetical protein |

|
 0.30106 0.30106
|