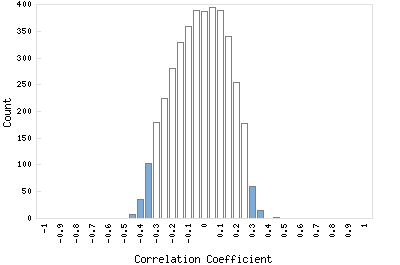
Correlation Catalog M. tuberculosis H37Rv: Rv2635 - hypothetical protein
Distribution of gene correlation: Rv2635 vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1309 | ATP synthase gamma chain atpG |

|
 -0.42916 -0.42916
| |
| Rv2257c | conserved hypothetical protein |

|
 -0.42372 -0.42372
| |
| Rv0721 | 30S ribosomal protein S5 rpsE |

|
 -0.42137 -0.42137
| |
| Rv0247c | succinate dehydrogenase iron-sulfur subunit |

|
 -0.41735 -0.41735
| |
| Rv1304 | ATP synthase A chain atpB |

|
 -0.4126 -0.4126
| |
| Rv3028c | electron transfer flavoprotein alpha subunit fixB |

|
 -0.40911 -0.40911
| |
| Rv1310 | ATP synthase beta chain atpD |

|
 -0.40803 -0.40803
| |
| Rv0722 | 50S ribosomal protein L30 rpmD |

|
 -0.40709 -0.40709
| |
| Rv3281 | conserved hypothetical protein |

|
 -0.39989 -0.39989
| |
| Rv1307 | ATP synthase delta chain atpH |

|
 -0.3975 -0.3975
| |
| Rv2245 | 3-oxoacyl-[acyl-carrier protein] synthase 1 kasA |


|
 -0.39614 -0.39614
| |
| Rv1438 | triosephosphate isomerase tpi |

|
 -0.39207 -0.39207
| |
| Rv0613c | hypothetical protein |

|
 -0.38921 -0.38921
| |
| Rv2102 | conserved hypothetical protein |

|
 -0.38772 -0.38772
| |
| Rv0733 | adenylate kinase adk |

|
 -0.38768 -0.38768
| |
| Rv1612 | tryptophan synthase, beta subunit trpB |

|
 -0.3876 -0.3876
| |
| Rv0642c | methoxy mycolic acid synthase 4 mmaA4 |

|
 -0.38523 -0.38523
| |
| Rv0157 | NAD(P) transhydrogenase beta subunit pntB |

|
 -0.38466 -0.38466
| |
| Rv1306 | ATP synthase B chain atpF |

|
 -0.38458 -0.38458
| |
| Rv3274c | acyl-CoA dehydrogenase fadE25 |

|
 -0.38446 -0.38446
| |
| Rv1078 | proline-rich antigen pra |

|
 -0.38351 -0.38351
| |
| Rv3799c | propionyl-CoA carboxylase beta chain 4 accD4 |

|
 -0.37585 -0.37585
| |
| Rv0248c | succinate dehydrogenase iron-sulfur subunit |

|
 -0.37495 -0.37495
| |
| Rv0734 | methionine aminopeptidase mapA |

|
 -0.37162 -0.37162
| |
| Rv0710 | 30S ribosomal protein S17 rpsQ |

|
 -0.37105 -0.37105
| |
| Rv3791 | short-chain type dehydrogenase/reductase |


|
 -0.37073 -0.37073
| |
| Rv3285 | bifunctional acetyl-/propionyl-coenzyme A carboxylase alpha chain accA3 |

|
 -0.37003 -0.37003
| |
| Rv0058 | replicative DNA helicase dnaB |

|
 -0.36979 -0.36979
| |
| Rv0227c | conserved membrane protein |

|
 -0.36971 -0.36971
| |
| Rv1677 | lipoprotein dsbF |


|
 -0.3689 -0.3689
| |
| Rv1311 | ATP synthase epsilon chain atpC |

|
 -0.36657 -0.36657
| |
| Rv2051c | polyprenol-monophosphomannose synthase ppm1 |

|
 -0.36476 -0.36476
| |
| Rv0760c | conserved hypothetical protein |

|
 -0.36444 -0.36444
| |
| Rv3801c | fatty-acid-CoA ligase fadD32 |


|
 -0.36391 -0.36391
| |
| Rv3318 | succinate dehydrogenase flavoprotein subunit sdhA |

|
 -0.3614 -0.3614
| |
| Rv3215 | isochorismate synthase entC |


|
 -0.36011 -0.36011
| |
| Rv0709 | 50S ribosomal protein L29 rpmC |

|
 -0.35693 -0.35693
| |
| Rv2855 | NADPH-dependent mycothiol reductase mtr |

|
 -0.35688 -0.35688
| |
| Rv2861c | methionine aminopeptidase mapB |

|
 -0.35604 -0.35604
| |
| Rv0632c | enoyl-CoA hydratase echA3 |

|
 -0.35576 -0.35576
| |
| Rv3800c | polyketide synthase pks13 |

|
 -0.35533 -0.35533
| |
| Rv3498c | MCE-family protein mce4B |

|
 -0.35491 -0.35491
| |
| Rv2215 | pyruvate dehydrogenase E2 component sucB |

|
 -0.35361 -0.35361
| |
| Rv2214c | short-chain type dehydrogenase ephD |


|
 -0.34898 -0.34898
| |
| Rv2244 | meromycolate extension acyl carrier protein acpM |


|
 -0.34772 -0.34772
| |
| Rv0479c | conserved membrane protein |

|
 -0.3477 -0.3477
| |
| Rv1836c | conserved hypothetical protein |

|
 -0.34767 -0.34767
| |
| Rv3011c | glutamyl-tRNA(gln) amidotransferase subunit A gatA |

|
 -0.3473 -0.3473
| |
| Rv1328 | glycogen phosphorylase glgP |

|
 -0.34716 -0.34716
| |
| Rv3024c | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase trmU |

|
 -0.34675 -0.34675
| |
| Rv0315 | beta-1,3-glucanase precursor |

|
 -0.34592 -0.34592
| |
| Rv2246 | 3-oxoacyl-[acyl-carrier protein] synthase 2 kasB |


|
 -0.3459 -0.3459
| |
| Rv3116 | molybdenum cofactor biosynthesis protein moeB2 |

|
 -0.34562 -0.34562
| |
| Rv2140c | conserved hypothetical protein |

|
 -0.34496 -0.34496
| |
| Rv2882c | ribosome recycling factor frr |

|
 -0.34413 -0.34413
| |
| Rv3030 | conserved hypothetical protein |


|
 -0.34384 -0.34384
| |
| Rv0954 | conserved membrane protein |

|
 -0.34357 -0.34357
| |
| Rv0183 | lysophospholipase |

|
 -0.34355 -0.34355
| |
| Rv0952 | succinyl-CoA synthetase alpha chain sucD |

|
 -0.34214 -0.34214
| |
| Rv3282 | conserved hypothetical protein |

|
 -0.34125 -0.34125
| |
| Rv2061c | conserved hypothetical protein |

|
 -0.34049 -0.34049
| |
| Rv3119 | molybdenum cofactor biosynthesis protein E moaE1 |

|
 -0.34043 -0.34043
| |
| Rv2859c | amidotransferase |

|
 -0.34003 -0.34003
| |
| Rv1610 | conserved membrane protein |

|
 -0.33778 -0.33778
| |
| Rv1630 | 30S ribosomal protein S1 rpsA |

|
 -0.33625 -0.33625
| |
| Rv0934 | periplasmic phosphate-binding lipoprotein pstS1 |

|
 -0.33609 -0.33609
| |
| Rv3528c | hypothetical protein |

|
 -0.33497 -0.33497
| |
| Rv0164 | conserved hypothetical protein |

|
 -0.33465 -0.33465
| |
| Rv1626 | two component system transcriptional regulator |

|
 -0.33449 -0.33449
| |
| Rv2220 | glutamine synthetase glnA1 |

|
 -0.33412 -0.33412
| |
| Rv0155 | NAD(P) transhydrogenase alpha subunit pntAa |

|
 -0.33403 -0.33403
| |
| Rv2970c | lipase/esterase lipN |

|
 -0.33358 -0.33358
| |
| Rv1097c | glycine and proline rich membrane protein |

|
 -0.33334 -0.33334
| |
| Rv3284 | conserved hypothetical protein |

|
 -0.33255 -0.33255
| |
| Rv1305 | ATP synthase C chain atpE |

|
 -0.33241 -0.33241
| |
| Rv3025c | cysteine desulfurase iscS |

|
 -0.3323 -0.3323
| |
| Rv0636 | conserved hypothetical protein |

|
 -0.33096 -0.33096
| |
| Rv0178 | MCE-associated membrane protein |

|
 -0.33007 -0.33007
| |
| Rv0464c | conserved hypothetical protein |

|
 -0.33002 -0.33002
| |
| Rv3598c | lysyl-tRNA synthetase 1 lysS |

|
 -0.32981 -0.32981
| |
| Rv3248c | adenosylhomocysteinase sahH |

|
 -0.3291 -0.3291
| |
| Rv2956 | conserved hypothetical protein |


|
 -0.32821 -0.32821
| |
| Rv2953 | conserved hypothetical protein |

|
 -0.32694 -0.32694
| |
| Rv3283 | thiosulfate sulfurtransferase sseA |

|
 -0.32659 -0.32659
| |
| Rv2604c | conserved hypothetical protein |

|
 -0.32625 -0.32625
| |
| Rv2427c | gamma-glutamyl phosphate reductase protein proA |

|
 -0.3262 -0.3262
| |
| Rv0637 | conserved hypothetical protein |

|
 -0.32585 -0.32585
| |
| Rv3852 | histone-like protein hns |

|
 -0.32436 -0.32436
| |
| Rv1919c | conserved hypothetical protein |

|
 -0.32433 -0.32433
| |
| Rv2845c | prolyl-tRNA synthetase proS |

|
 -0.32372 -0.32372
| |
| Rv2453c | molybdopterin-guanine dinucleotide biosynthesis protein A mobA |

|
 -0.32233 -0.32233
| |
| Rv1837c | malate synthase G glcB |

|
 -0.3223 -0.3223
| |
| Rv2394 | gamma-glutamyltranspeptidase precursor ggtB |

|
 -0.3215 -0.3215
| |
| Rv1437 | phosphoglycerate kinase pgk |

|
 -0.32115 -0.32115
| |
| Rv3438 | conserved hypothetical protein |

|
 -0.32099 -0.32099
| |
| Rv0177 | MCE-associated protein |

|
 -0.32079 -0.32079
| |
| Rv3029c | electron transfer flavoprotein beta subunit fixA |

|
 -0.32064 -0.32064
| |
| Rv3240c | preprotein translocase secA1 |

|
 -0.32045 -0.32045
| |
| Rv3151 | NADH dehydrogenase I chain G nuoG |

|
 -0.31899 -0.31899
| |
| Rv0685 | iron-regulated elongation factor tu tuf |


|
 -0.31892 -0.31892
| |
| Rv1387 | PPE family protein |

|
 -0.31871 -0.31871
| |
| Rv0462 | dihydrolipoamide dehydrogenase lpd |

|
 -0.31871 -0.31871
| |
| Rv2476c | NAD-dependent glutamate dehydrogenase gdh |

|
 -0.31775 -0.31775
| |
| Rv2477c | macrolide-transport ATP-binding protein ABC transporter |

|
 -0.31681 -0.31681
| |
| Rv1023 | enolase eno |

|
 -0.31592 -0.31592
| |
| Rv2243 | malonyl CoA-acyl carrier protein transacylase fabD |

|
 -0.31574 -0.31574
| |
| Rv2200c | transmembrane cytochrome C oxidase subunit II ctaC |

|
 -0.31556 -0.31556
| |
| Rv0761c | zinc-type alcohol dehydrogenase NAD dependent adhB |

|
 -0.31502 -0.31502
| |
| Rv2195 | Rieske iron-sulfur protein qcrA |

|
 -0.31449 -0.31449
| |
| Rv1703c | catechol-O-methyltransferase |


|
 -0.31396 -0.31396
| |
| Rv1447c | glucose-6-phosphate 1-dehydrogenase zwf2 |

|
 -0.31301 -0.31301
| |
| Rv3458c | 30S ribosomal protein S4 rpsD |

|
 -0.31298 -0.31298
| |
| Rv2376c | low molecular weight protein antigen cfp2 |

|
 -0.31249 -0.31249
| |
| Rv3682 | bifunctional membrane-associated penicillin-binding protein 1A/1B ponA2 |

|
 -0.3119 -0.3119
| |
| Rv0005 | DNA gyrase subunit B gyrB |

|
 -0.31167 -0.31167
| |
| Rv1770 | conserved hypothetical protein |

|
 -0.31152 -0.31152
| |
| Rv2228c | conserved hypothetical protein |

|
 -0.31117 -0.31117
| |
| Rv3804c | secreted fibronectin-binding protein antigen fbpA |

|
 -0.31055 -0.31055
| |
| Rv1312 | conserved secreted protein |

|
 -0.31022 -0.31022
| |
| Rv1508c | membrane protein |

|
 -0.31003 -0.31003
| |
| Rv0423c | thiamine biosynthesis protein thiC |

|
 -0.30994 -0.30994
| |
| Rv0708 | 50S ribosomal protein L16 rplP |

|
 -0.30834 -0.30834
| |
| Rv1886c | secreted fibronectin-binding protein antigen 85-B fbpB |

|
 -0.30807 -0.30807
| |
| Rv2524c | fatty-acid synthase fas |

|
 -0.30776 -0.30776
| |
| Rv3417c | 60 kda chaperonin 1 groEL1 |

|
 -0.30775 -0.30775
| |
| Rv3780 | conserved hypothetical protein |

|
 -0.30771 -0.30771
| |
| Rv2082 | conserved hypothetical protein |

|
 -0.30742 -0.30742
| |
| Rv3150 | NADH dehydrogenase I chain F nuoF |

|
 -0.30733 -0.30733
| |
| Rv1038c | Esat-6 like protein esxJ |

|
 -0.30719 -0.30719
| |
| Rv1611 | indole-3-glycerol phosphate synthase trpC |

|
 -0.307 -0.307
| |
| Rv0433 | conserved hypothetical protein |

|
 -0.30685 -0.30685
| |
| Rv0667 | DNA-directed RNA polymerase beta chain rpoB |

|
 -0.30639 -0.30639
| |
| Rv2587c | protein-export membrane protein secD |

|
 -0.30545 -0.30545
| |
| Rv2981c | D-alanine-D-alanine ligase ddlA |

|
 -0.30428 -0.30428
| |
| Rv1172c | PE family protein |

|
 -0.30418 -0.30418
| |
| Rv1614 | prolipoprotein diacylglyceryl transferases lgt |

|
 -0.30413 -0.30413
| |
| Rv0684 | elongation factor G fusA1 |

|
 -0.30374 -0.30374
| |
| Rv0822c | conserved hypothetical protein |

|
 -0.30306 -0.30306
| |
| Rv3153 | NADH dehydrogenase I chain I nuoI |

|
 -0.30211 -0.30211
| |
| Rv3496c | MCE-family protein mce4D |

|
 -0.30206 -0.30206
| |
| Rv3818 | hypothetical protein |

|
 -0.3019 -0.3019
| |
| Rv3418c | 10 kda chaperonin groES |

|
 -0.30155 -0.30155
| |
| Rv3648c | cold shock protein A cspA |

|
 -0.30118 -0.30118
| |
| Rv3043c | cytochrome C oxidase polypeptide I ctaD |

|
 -0.3003 -0.3003
| |
| Rv3158 | NADH dehydrogenase I chain N nuoN |

|
 -0.30008 -0.30008
| |
| Rv2881c | membrane phosphatidate cytidylyltransferase cdsA |

|
 -0.30005 -0.30005
|
Genes with positive correlation 
Categories Enriched  : K
: K
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2631 | conserved hypothetical protein |

|
 0.46828 0.46828
| |
| Rv2639c | conserved membrane protein |

|
 0.45309 0.45309
| |
| Rv2323c | conserved hypothetical protein |

|
 0.39486 0.39486
| |
| Rv0767c | conserved hypothetical protein |

|
 0.38776 0.38776
| |
| Rv1052 | hypothetical protein |

|
 0.38656 0.38656
| |
| Rv0375c | carbon monoxyde dehydrogenase medium chain |

|
 0.37639 0.37639
| |
| Rv0077c | oxidoreductase |

|
 0.37043 0.37043
| |
| Rv2656c | phiRv2 phage protein |

|
 0.36888 0.36888
| |
| Rv0232 | transcriptional regulator, tetR/acrR-family |

|
 0.36465 0.36465
| |
| Rv1503c | conserved hypothetical protein |

|
 0.36449 0.36449
| |
| Rv1946c | lipoprotein lppG |

|
 0.36333 0.36333
| |
| Rv2662 | hypothetical protein |

|
 0.36226 0.36226
| |
| Rv0656c | conserved hypothetical protein |

|
 0.35893 0.35893
| |
| Rv2615c | PE-PGRS family protein |

|
 0.35733 0.35733
| |
| Rv0335c | PE family protein |

|
 0.35367 0.35367
| |
| Rv2516c | hypothetical protein |

|
 0.35244 0.35244
| |
| Rv1861 | conserved membrane protein |

|
 0.35031 0.35031
| |
| Rv0259c | conserved hypothetical protein |

|
 0.34714 0.34714
| |
| Rv2517c | hypothetical protein |

|
 0.34335 0.34335
| |
| Rv0998 | conserved hypothetical protein |

|
 0.34051 0.34051
| |
| Rv3288c | hypothetical protein usfY |

|
 0.34032 0.34032
| |
| Rv2661c | hypothetical protein |

|
 0.33833 0.33833
| |
| Rv3018c | PPE family protein |

|
 0.33796 0.33796
| |
| Rv2657c | phiRv2 phage protein |

|
 0.33772 0.33772
| |
| Rv0027 | hypothetical protein |

|
 0.33687 0.33687
| |
| Rv3333c | hypothetical proline rich protein |

|
 0.33492 0.33492
| |
| Rv2827c | hypothetical protein |

|
 0.33453 0.33453
| |
| Rv2514c | conserved hypothetical protein |

|
 0.33425 0.33425
| |
| Rv0122 | hypothetical protein |

|
 0.33391 0.33391
| |
| Rv0328 | transcriptional regulator, tetR/acrR-family |

|
 0.33136 0.33136
| |
| Rv2269c | hypothetical protein |

|
 0.3313 0.3313
| |
| Rv2663 | hypothetical protein |

|
 0.33095 0.33095
| |
| Rv1051c | conserved hypothetical protein |

|
 0.3271 0.3271
| |
| Rv0355c | PPE family protein |

|
 0.3264 0.3264
| |
| Rv3383c | polyprenyl synthetase idsB |

|
 0.32466 0.32466
| |
| Rv2643 | arsenic-transport membrane protein arsC |

|
 0.32364 0.32364
| |
| Rv1150 | transposase |

|
 0.32329 0.32329
| |
| Rv0327c | cytochrome P450 135A1 cyp135A1 |

|
 0.32327 0.32327
| |
| Rv3129 | conserved hypothetical protein |

|
 0.32304 0.32304
| |
| Rv2638 | conserved hypothetical protein |

|
 0.32298 0.32298
| |
| Rv2669 | conserved hypothetical protein |


|
 0.32251 0.32251
| |
| Rv2655c | phiRv2 phage protein |

|
 0.32152 0.32152
| |
| Rv1804c | conserved hypothetical protein |

|
 0.32108 0.32108
| |
| Rv1075c | conserved exported protein |

|
 0.32035 0.32035
| |
| Rv0264c | conserved hypothetical protein |

|
 0.31978 0.31978
| |
| Rv0137c | peptide methionine sulfoxide reductase msrA |

|
 0.31887 0.31887
| |
| Rv3188 | conserved hypothetical protein |

|
 0.31805 0.31805
| |
| Rv3180c | hypothetical alanine rich protein |

|
 0.31794 0.31794
| |
| Rv1000 |

|
 0.3165 0.3165
| ||
| Rv3833 | transcriptional regulator, araC-family |

|
 0.31636 0.31636
| |
| Rv0735 | alternative RNA polymerase sigma factor sigL |

|
 0.31629 0.31629
| |
| Rv1913 | conserved hypothetical protein |

|
 0.31612 0.31612
| |
| Rv0325 | hypothetical protein |

|
 0.31533 0.31533
| |
| Rv1733c | conserved membrane protein |

|
 0.31441 0.31441
| |
| Rv0331 | dehydrogenase/reductase |

|
 0.31431 0.31431
| |
| Rv3845 | hypothetical protein |

|
 0.31344 0.31344
| |
| Rv2660c | hypothetical protein |

|
 0.31211 0.31211
| |
| Rv2874 | cytochrome C biogenesis protein dipZ |

|
 0.31083 0.31083
| |
| Rv3290c | L-lysine-epsilon aminotransferase lat |

|
 0.31039 0.31039
| |
| Rv1141c | enoyl-CoA hydratase echA11 |

|
 0.30938 0.30938
| |
| Rv3347c | PPE family protein |

|
 0.30824 0.30824
| |
| Rv1131 | citrate synthase I gltA1 |

|
 0.30813 0.30813
| |
| Rv3125c | PPE family protein |

|
 0.3069 0.3069
| |
| Rv2295 | conserved hypothetical protein |

|
 0.30652 0.30652
| |
| Rv2034 | arsR-type repressor protein |

|
 0.30647 0.30647
| |
| Rv0648 | alpha-mannosidase |

|
 0.30594 0.30594
| |
| Rv3359 | oxidoreductase |

|
 0.30551 0.30551
| |
| Rv2014 | transposase |

|
 0.30512 0.30512
| |
| Rv3334 | transcriptional regulator, merR-family |

|
 0.30485 0.30485
| |
| Rv1574 | phiRv1 phage protein |

|
 0.30398 0.30398
| |
| Rv3182 | conserved hypothetical protein |

|
 0.30263 0.30263
| |
| Rv2644c | hypothetical protein |

|
 0.30237 0.30237
| |
| Rv2089c | dipeptidase pepE |

|
 0.30135 0.30135
| |
| Rv2541 | hypothetical alanine rich protein |

|
 0.3013 0.3013
| |
| Rv3425 | PPE family protein |

|
 0.30124 0.30124
| |
| Rv0307c | hypothetical protein |

|
 0.30024 0.30024
| |
| Rv3911 | alternative RNA polymerase sigma factor sigM |

|
 0.30014 0.30014
|