Correlation Catalog M. tuberculosis H37Rv: Rv2377c - hypothetical protein mbtH
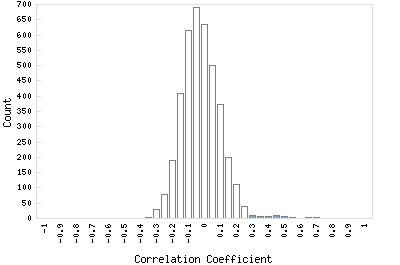
Distribution of gene correlation: Rv2377c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3841 | bacterioferritin bfrB |

|
 -0.46026 -0.46026
| |
| Rv1876 | bacterioferritin bfrA |

|
 -0.41199 -0.41199
| |
| Rv2284 | esterase lipM |

|
 -0.36179 -0.36179
| |
| Rv1988 | methyltransferase |

|
 -0.30677 -0.30677
| |
| Rv2019 | hypothetical protein |

|
 -0.30171 -0.30171
| |
| Rv1475c | iron-regulated aconitate hydratase acn |

|
 -0.30097 -0.30097
|
Genes with positive correlation 
Categories Enriched  : Q
: Q
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2381c | polyketide synthetase mbtD |

|
 0.75929 0.75929
| |
| Rv2380c | peptide synthetase mbtE |

|
 0.73539 0.73539
| |
| Rv2383c | phenyloxazoline synthase mbtB |

|
 0.72187 0.72187
| |
| Rv2379c | peptide synthetase mbtF |

|
 0.66922 0.66922
| |
| Rv2382c | polyketide synthetase mbtC |

|
 0.66869 0.66869
| |
| Rv3839 | conserved hypothetical protein |

|
 0.57949 0.57949
| |
| Rv0285 | PE family protein |

|
 0.5697 0.5697
| |
| Rv0282 | conserved hypothetical protein |

|
 0.55214 0.55214
| |
| Rv0286 | PPE family protein |

|
 0.54763 0.54763
| |
| Rv0283 | conserved membrane protein |

|
 0.54076 0.54076
| |
| Rv0289 | conserved hypothetical protein |

|
 0.53381 0.53381
| |
| Rv3402c | conserved hypothetical protein |

|
 0.53042 0.53042
| |
| Rv1347c | conserved hypothetical protein |

|
 0.51963 0.51963
| |
| Rv0287 | esat-6 like protein esxG |

|
 0.51844 0.51844
| |
| Rv1463 | ABC transporter ATP-binding protein |

|
 0.51424 0.51424
| |
| Rv0288 | low molecular weight protein antigen 7 esxH |

|
 0.51101 0.51101
| |
| Rv0284 | conserved membrane protein |

|
 0.49892 0.49892
| |
| Rv3020c | Esat-6 like protein esxS |

|
 0.49743 0.49743
| |
| Rv2123 | PPE family protein |

|
 0.49353 0.49353
| |
| Rv3019c | Esat-6 like protein esxR |

|
 0.49271 0.49271
| |
| Rv1349 | drugs-transport transmembrane ATP-binding protein ABC transporter |

|
 0.4839 0.4839
| |
| Rv2386c | isochorismate synthase mbtI |


|
 0.47215 0.47215
| |
| Rv0451c | membrane protein mmpS4 |

|
 0.47016 0.47016
| |
| Rv0465c | transcriptional regulator |

|
 0.46903 0.46903
| |
| Rv3403c | hypothetical protein |

|
 0.4686 0.4686
| |
| Rv2378c | lysine-N-oxygenase mbtG |

|
 0.46685 0.46685
| |
| Rv1461 | conserved hypothetical protein |

|
 0.46669 0.46669
| |
| Rv0290 | conserved membrane protein |

|
 0.44948 0.44948
| |
| Rv1466 | conserved hypothetical protein |

|
 0.44836 0.44836
| |
| Rv3017c | Esat-6 like protein esxQ |

|
 0.44736 0.44736
| |
| Rv1464 | cysteine desulfurase csd |

|
 0.43804 0.43804
| |
| Rv0292 | conserved membrane protein |

|
 0.42827 0.42827
| |
| Rv1346 | acyl-CoA dehydrogenase fadE14 |

|
 0.41695 0.41695
| |
| Rv1465 | nitrogen fixation related protein |

|
 0.40865 0.40865
| |
| Rv3840 | transcriptional regulator |

|
 0.40141 0.40141
| |
| Rv1462 | conserved hypothetical protein |

|
 0.39938 0.39938
| |
| Rv1344 | acyl carrier protein |


|
 0.39129 0.39129
| |
| Rv1005c | para-aminobenzoate synthase component I pabD |


|
 0.38169 0.38169
| |
| Rv0464c | conserved hypothetical protein |

|
 0.37659 0.37659
| |
| Rv1343c | lipoprotein lprD |

|
 0.35498 0.35498
| |
| Rv1348 | drugs-transport transmembrane ATP-binding protein ABC transporter |

|
 0.33644 0.33644
| |
| Rv0291 | membrane-anchored mycosin mycP3 |

|
 0.33505 0.33505
| |
| Rv2933 | phenolpthiocerol synthesis type-I polyketide synthase ppsC |

|
 0.33226 0.33226
| |
| Rv0005 | DNA gyrase subunit B gyrB |

|
 0.32624 0.32624
| |
| Rv1345 | polyketide synthase fadD33 |


|
 0.32129 0.32129
| |
| Rv3693 | conserved membrane protein |

|
 0.31852 0.31852
| |
| Rv1596 | nicotinate-nucleotide pyrophosphatase nadC |

|
 0.31049 0.31049
| |
| Rv2967c | pyruvate carboxylase pca |

|
 0.30962 0.30962
| |
| Rv2859c | amidotransferase |

|
 0.30599 0.30599
| |
| Rv2109c | proteasome alpha subunit prcA |

|
 0.30513 0.30513
|