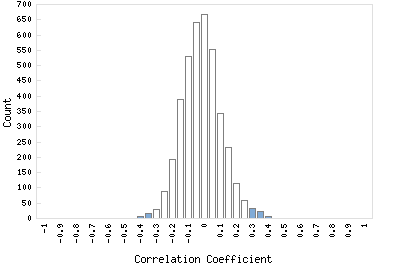
Correlation Catalog M. tuberculosis H37Rv: Rv2108 - PPE family protein
Distribution of gene correlation: Rv2108 vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3295 | transcriptional regulator, tetR-family |

|
 -0.45174 -0.45174
| |
| Rv0655 | ribonucleotide-transport ATP-binding protein ABC transporter mkl |

|
 -0.42214 -0.42214
| |
| Rv0171 | MCE-family protein mce1C |

|
 -0.38237 -0.38237
| |
| Rv0170 | MCE-family protein mce1B |

|
 -0.38181 -0.38181
| |
| Rv0169 | MCE-family protein mce1A |

|
 -0.37613 -0.37613
| |
| Rv0166 | fatty-acid-CoA ligase fadD5 |


|
 -0.37054 -0.37054
| |
| Rv3083 | monooxygenase |

|
 -0.36077 -0.36077
| |
| Rv0995 | ribosomal-protein-alanine acetyltransferase rimJ |

|
 -0.34877 -0.34877
| |
| Rv0172 | MCE-family protein mce1D |

|
 -0.34539 -0.34539
| |
| Rv0167 | hypothetical membrane protein yrbE1A |

|
 -0.34346 -0.34346
| |
| Rv0250c | conserved hypothetical protein |

|
 -0.34172 -0.34172
| |
| Rv2406c | conserved hypothetical protein |

|
 -0.33581 -0.33581
| |
| Rv2485c | carboxylesterase lipQ |

|
 -0.3296 -0.3296
| |
| Rv3084 | acetyl-hydrolase/esterase lipR |

|
 -0.32885 -0.32885
| |
| Rv0168 | hypothetical membrane protein yrbE1B |

|
 -0.32398 -0.32398
| |
| Rv3146 | NADH dehydrogenase I chain B nuoB |

|
 -0.32277 -0.32277
| |
| Rv3147 | NADH dehydrogenase I chain C nuoC |

|
 -0.31894 -0.31894
| |
| Rv0458 | aldehyde dehydrogenase |

|
 -0.30895 -0.30895
| |
| Rv0242c | 3-oxoacyl-[acyl-carrier protein] reductase fabG4 |


|
 -0.3062 -0.3062
| |
| Rv2681 | conserved alanine rich protein |

|
 -0.30557 -0.30557
| |
| Rv0174 | MCE-family protein mce1F |

|
 -0.30365 -0.30365
| |
| Rv3086 | zinc-type alcohol dehydrogenase adhD |

|
 -0.30309 -0.30309
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2107 | PE family protein |

|
 0.54507 0.54507
| |
| Rv0474 | transcriptional regulator |

|
 0.46024 0.46024
| |
| Rv0244c | acyl-CoA dehydrogenase fadE5 |

|
 0.44187 0.44187
| |
| Rv3484 | hypothetical protein cpsA |

|
 0.43496 0.43496
| |
| Rv0475 | iron-regulated heparin binding hemagglutinin hbhA |

|
 0.42344 0.42344
| |
| Rv1779c | hypothetical membrane protein |

|
 0.41893 0.41893
| |
| Rv3252c | transmembrane alkane 1-monooxygenase alkB |

|
 0.41497 0.41497
| |
| Rv2428 | alkyl hydroperoxide reductase C protein ahpC |

|
 0.41358 0.41358
| |
| Rv2930 | fatty-acid-CoA ligase fadD26 |


|
 0.40014 0.40014
| |
| Rv0726c | conserved hypothetical protein |

|
 0.39638 0.39638
| |
| Rv3251c | rubredoxin rubA |

|
 0.39527 0.39527
| |
| Rv0469 | mycolic acid synthase umaA |

|
 0.39346 0.39346
| |
| Rv0467 | isocitrate lyase icl |

|
 0.39298 0.39298
| |
| Rv0476 | conserved membrane protein |

|
 0.38984 0.38984
| |
| Rv1168c | PPE family protein |

|
 0.38769 0.38769
| |
| Rv2518c | lipoprotein lppS |

|
 0.38372 0.38372
| |
| Rv2784c | lipoprotein lppU |

|
 0.37689 0.37689
| |
| Rv0886 | NADPH:adrenodoxin oxidoreductase fprB |

|
 0.37289 0.37289
| |
| Rv2429 | alkyl hydroperoxide reductase D protein ahpD |

|
 0.37059 0.37059
| |
| Rv1169c | PE family protein |

|
 0.37008 0.37008
| |
| Rv3515c | fatty-acid-CoA ligase fadD19 |


|
 0.36568 0.36568
| |
| Rv0477 | conserved secreted protein |

|
 0.36309 0.36309
| |
| Rv1636 | iron-regulated conserved hypothetical protein |

|
 0.36173 0.36173
| |
| Rv2724c | acyl-CoA dehydrogenase fadE20 |

|
 0.36117 0.36117
| |
| Rv3249c | transcriptional regulator, tetR-family |

|
 0.35999 0.35999
| |
| Rv1647 | conserved hypothetical protein |

|
 0.35834 0.35834
| |
| Rv1592c | conserved hypothetical protein |

|
 0.35808 0.35808
| |
| Rv0711 | arylsulfatase atsA |

|
 0.35589 0.35589
| |
| Rv1839c | conserved hypothetical protein |

|
 0.354 0.354
| |
| Rv0145 | conserved hypothetical protein |

|
 0.35092 0.35092
| |
| Rv3516 | enoyl-CoA hydratase echA19 |

|
 0.3506 0.3506
| |
| Rv0468 | 3-hydroxybutyryl-CoA dehydrogenase fadB2 |

|
 0.34716 0.34716
| |
| Rv1534 | transcriptional regulator |

|
 0.34624 0.34624
| |
| Rv3522 | lipid transfer protein or keto acyl-CoA thiolase ltp4 |

|
 0.34404 0.34404
| |
| Rv0146 | conserved hypothetical protein |

|
 0.34369 0.34369
| |
| Rv2452c | hypothetical protein |

|
 0.34098 0.34098
| |
| Rv2226 | conserved hypothetical protein |

|
 0.33991 0.33991
| |
| Rv0859 | acyl-CoA thiolase fadA |

|
 0.33591 0.33591
| |
| Rv1533 | conserved hypothetical protein |

|
 0.33391 0.33391
| |
| Rv2640c | transcriptional regulator, arsR-family |

|
 0.33313 0.33313
| |
| Rv2959c | methyltransferase/methylase |

|
 0.32834 0.32834
| |
| Rv0352 | chaperone protein dnaJ1 |

|
 0.32129 0.32129
| |
| Rv3645 | conserved membrane protein |

|
 0.32046 0.32046
| |
| Rv1292 | arginyl-tRNA synthetase argS |

|
 0.31938 0.31938
| |
| Rv0940c | oxidoreductase |

|
 0.3191 0.3191
| |
| Rv3250c | rubredoxin rubB |

|
 0.31828 0.31828
| |
| Rv0350 | chaperone protein dnaK |

|
 0.31824 0.31824
| |
| Rv0211 | iron-regulated phosphoenolpyruvate carboxykinase pckA |

|
 0.31816 0.31816
| |
| Rv1811 | Mg2+ transport ATPase C mgtC |

|
 0.31658 0.31658
| |
| Rv2705c | conserved hypothetical protein |

|
 0.3163 0.3163
| |
| Rv1532c | conserved hypothetical protein |

|
 0.31351 0.31351
| |
| Rv1015c | 50S ribosomal protein L25 rplY |

|
 0.31217 0.31217
| |
| Rv2707 | conserved alanine and leucine rich membrane protein |

|
 0.31186 0.31186
| |
| Rv0068 | oxidoreductase |


|
 0.31091 0.31091
| |
| Rv1194c | conserved hypothetical protein |


|
 0.31065 0.31065
| |
| Rv3406 | dioxygenase |

|
 0.30964 0.30964
| |
| Rv0885 | conserved hypothetical protein |

|
 0.30821 0.30821
| |
| Rv3901c | membrane protein |

|
 0.3077 0.3077
| |
| Rv3601c | aspartate 1-decarboxylase precursor panD |

|
 0.30714 0.30714
| |
| Rv3602c | pantoate-beta-alanine ligase panC |

|
 0.30483 0.30483
| |
| Rv0905 | enoyl-CoA hydratase echA6 |

|
 0.30474 0.30474
| |
| Rv1461 | conserved hypothetical protein |

|
 0.30402 0.30402
| |
| Rv0351 | chaperone grpE |

|
 0.30379 0.30379
| |
| Rv3574 | transcriptional regulator, tetR-family |

|
 0.30189 0.30189
|