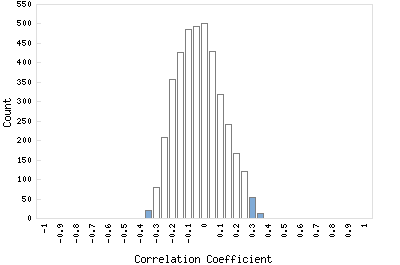
Correlation Catalog M. tuberculosis H37Rv: Rv2070c - precorrin-6x reductase cobK
Distribution of gene correlation: Rv2070c vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3261 | F420 biosynthesis protein fbiA |

|
 -0.36561 -0.36561
| |
| Rv3265c | dTDP-rha:a-D-glcnac-diphosphoryl polyprenol, a-3-L-rhamnosyl transferase wbbL1 |

|
 -0.34399 -0.34399
| |
| Rv3709c | aspartokinase ask |

|
 -0.33407 -0.33407
| |
| Rv0641 | 50S ribosomal protein L1 rplA |

|
 -0.33208 -0.33208
| |
| Rv1015c | 50S ribosomal protein L25 rplY |

|
 -0.32791 -0.32791
| |
| Rv2487c | PE-PGRS family protein |

|
 -0.3263 -0.3263
| |
| Rv0020c | conserved hypothetical protein |

|
 -0.31596 -0.31596
| |
| Rv2993c | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase |

|
 -0.31377 -0.31377
| |
| Rv3325 | transposase |

|
 -0.31178 -0.31178
| |
| Rv1483 | 3-oxoacyl-[acyl-carrier protein] reductase fabG1 |


|
 -0.3107 -0.3107
| |
| Rv0638 | preprotein translocase secE1 |

|
 -0.31024 -0.31024
| |
| Rv1859 | molybdenum-transport ATP-binding protein ABC transporter modC |

|
 -0.30989 -0.30989
| |
| Rv0299 | hypothetical protein |

|
 -0.30958 -0.30958
| |
| Rv3272 | conserved hypothetical protein |

|
 -0.30913 -0.30913
| |
| Rv3116 | molybdenum cofactor biosynthesis protein moeB2 |

|
 -0.30792 -0.30792
| |
| Rv2871 | conserved hypothetical protein |

|
 -0.30775 -0.30775
| |
| Rv1451 | cytochrome C oxidase assembly factor ctaB |

|
 -0.30525 -0.30525
| |
| Rv3276c | phosphoribosylaminoimidazole carboxylase ATPase subunit purK |

|
 -0.30388 -0.30388
| |
| Rv3381c | transposase |

|
 -0.3031 -0.3031
| |
| Rv0023 | transcriptional regulator |

|
 -0.30206 -0.30206
| |
| Rv2697c | deoxyuridine 5-triphosphate nucleotidohydrolase dut |

|
 -0.30201 -0.30201
| |
| Rv2117 | conserved hypothetical protein |

|
 -0.30174 -0.30174
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0104 | conserved hypothetical protein |

|
 0.41222 0.41222
| |
| Rv1714 | oxidoreductase |


|
 0.3985 0.3985
| |
| Rv2388c | oxygen-independent coproporphyrinogen III oxidase hemN |

|
 0.39702 0.39702
| |
| Rv0130 | conserved hypothetical protein |

|
 0.39261 0.39261
| |
| Rv2408 | PE family protein |

|
 0.38935 0.38935
| |
| Rv0107c | cation-transporter ATPase I ctpI |

|
 0.38814 0.38814
| |
| Rv0120c | elongation factor G fusA2 |

|
 0.38658 0.38658
| |
| Rv1004c | membrane protein |

|
 0.36958 0.36958
| |
| Rv1058 | medium chain fatty-acid-CoA ligase fadD14 |


|
 0.36603 0.36603
| |
| Rv2419c | phosphoglycerate mutase |

|
 0.36137 0.36137
| |
| Rv1330c | conserved hypothetical protein |

|
 0.35843 0.35843
| |
| Rv0370c | oxidoreductase |

|
 0.35767 0.35767
| |
| Rv1752 | conserved hypothetical protein |

|
 0.35542 0.35542
| |
| Rv2262c | conserved hypothetical protein |

|
 0.35217 0.35217
| |
| Rv0674 | conserved hypothetical protein |

|
 0.35215 0.35215
| |
| Rv0380c | RNA methyltransferase |

|
 0.3493 0.3493
| |
| Rv3634c | UDP-glucose 4-epimerase galE1 |


|
 0.34755 0.34755
| |
| Rv0976c | conserved hypothetical protein |

|
 0.34736 0.34736
| |
| Rv0372c | conserved hypothetical protein |

|
 0.34675 0.34675
| |
| Rv0334 | alpha-D-glucose-1-phosphate thymidylyltransferase rmlA |

|
 0.34634 0.34634
| |
| Rv0974c | acetyl-/propionyl-CoA carboxylase beta subunit accD2 |

|
 0.34567 0.34567
| |
| Rv1140 | membrane protein |

|
 0.34275 0.34275
| |
| Rv0634c | glyoxalase II |

|
 0.34226 0.34226
| |
| Rv1972 | MCE-associated membrane protein |

|
 0.33888 0.33888
| |
| Rv0506 | membrane protein mmpS2 |

|
 0.33873 0.33873
| |
| Rv0052 | conserved hypothetical protein |

|
 0.33761 0.33761
| |
| Rv1278 | hypothetical protein |

|
 0.33676 0.33676
| |
| Rv3776 | conserved hypothetical protein |

|
 0.33555 0.33555
| |
| Rv1551 | acyltransferase plsB1 |

|
 0.33402 0.33402
| |
| Rv3614c | conserved hypothetical protein |

|
 0.33354 0.33354
| |
| Rv1777 | cytochrome P450 144 cyp144 |

|
 0.33334 0.33334
| |
| Rv1996 | conserved hypothetical protein |

|
 0.33255 0.33255
| |
| Rv1880c | cytochrome P450 140 cyp140 |

|
 0.33219 0.33219
| |
| Rv2090 | 5-3 exonuclease |

|
 0.3304 0.3304
| |
| Rv1710 | conserved hypothetical protein |

|
 0.3279 0.3279
| |
| Rv3109 | molybdenum cofactor biosynthesis protein A moaA1 |

|
 0.32646 0.32646
| |
| Rv1514c | conserved hypothetical protein |

|
 0.32396 0.32396
| |
| Rv3809c | UDP-galactopyranose mutase glf |

|
 0.32311 0.32311
| |
| Rv1393c | monooxygenase |

|
 0.32247 0.32247
| |
| Rv0508 | conserved hypothetical protein |


|
 0.31931 0.31931
| |
| Rv2823c | conserved hypothetical protein |

|
 0.31898 0.31898
| |
| Rv2416c | enhanced intracellular survival protein eis |


|
 0.3188 0.3188
| |
| Rv3366 | tRNA/rRNA methylase spoU |

|
 0.31868 0.31868
| |
| Rv1639c | conserved membrane protein |

|
 0.31785 0.31785
| |
| Rv2325c | conserved hypothetical protein |

|
 0.31748 0.31748
| |
| Rv1544 | ketoacyl reductase |

|
 0.31679 0.31679
| |
| Rv0449c | conserved hypothetical protein |

|
 0.31666 0.31666
| |
| Rv0150c | conserved hypothetical protein |

|
 0.3165 0.3165
| |
| Rv0941c | conserved hypothetical protein |

|
 0.31615 0.31615
| |
| Rv0102 | conserved membrane protein |

|
 0.31462 0.31462
| |
| Rv2260 | conserved hypothetical protein |

|
 0.31446 0.31446
| |
| Rv1582c | phiRv1 phage protein |

|
 0.31424 0.31424
| |
| Rv0261c | membrane nitrite extrusion protein narK3 |

|
 0.3136 0.3136
| |
| Rv0924c | divalent cation-transport membrane protein mntH |

|
 0.31084 0.31084
| |
| Rv1530 | alcohol dehydrogenase adh |


|
 0.30978 0.30978
| |
| Rv1414 | conserved hypothetical protein |

|
 0.30911 0.30911
| |
| Rv1112 | GTP binding protein |

|
 0.3085 0.3085
| |
| Rv1138c | oxidoreductase |

|
 0.30702 0.30702
| |
| Rv2946c | polyketide synthase pks1 |

|
 0.30634 0.30634
| |
| Rv3615c | conserved hypothetical protein |

|
 0.30615 0.30615
| |
| Rv1575 | phiRv1 phage protein |

|
 0.30512 0.30512
| |
| Rv0798c | 29 kda antigen cfp29 |

|
 0.30509 0.30509
| |
| Rv1029 | potassium-transporting ATPase A chain kdpA |

|
 0.30502 0.30502
| |
| Rv1634 | drug efflux membrane protein |




|
 0.30418 0.30418
| |
| Rv1738 | conserved hypothetical protein |

|
 0.30301 0.30301
| |
| Rv0074 | conserved hypothetical protein |

|
 0.30294 0.30294
| |
| Rv0304c | PPE family protein |

|
 0.30254 0.30254
| |
| Rv2240c | hypothetical protein |

|
 0.30195 0.30195
| |
| Rv2024c | conserved hypothetical protein |

|
 0.30191 0.30191
|