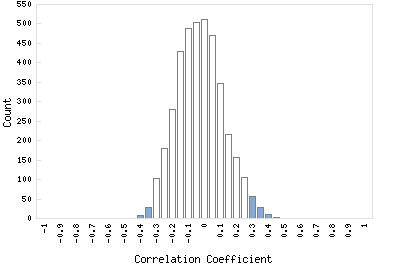
Correlation Catalog M. tuberculosis H37Rv: Rv1924c - hypothetical protein
Distribution of gene correlation: Rv1924c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3017c | Esat-6 like protein esxQ |

|
 -0.40352 -0.40352
| |
| Rv2126c | PE-PGRS family protein |

|
 -0.39453 -0.39453
| |
| Rv1842c | conserved membrane protein |

|
 -0.39028 -0.39028
| |
| Rv3653 | PE-PGRS family protein |

|
 -0.37394 -0.37394
| |
| Rv1361c | PPE family protein |

|
 -0.37212 -0.37212
| |
| Rv2853 | PE-PGRS family protein |

|
 -0.37193 -0.37193
| |
| Rv1295 | threonine synthase thrC |

|
 -0.36743 -0.36743
| |
| Rv1837c | malate synthase G glcB |

|
 -0.36733 -0.36733
| |
| Rv0901 | hypothetical exported protein |

|
 -0.35682 -0.35682
| |
| Rv2553c | conserved membrane protein |

|
 -0.34961 -0.34961
| |
| Rv0351 | chaperone grpE |

|
 -0.34729 -0.34729
| |
| Rv1396c | PE-PGRS family protein |

|
 -0.34655 -0.34655
| |
| Rv2490c | PE-PGRS family protein |

|
 -0.34426 -0.34426
| |
| Rv1091 | PE-PGRS family protein |

|
 -0.34303 -0.34303
| |
| Rv3905c | Esat-6 like protein esxF |

|
 -0.34013 -0.34013
| |
| Rv3167c | transcriptional regulator, tetR-family |

|
 -0.33927 -0.33927
| |
| Rv3673c | membrane-anchored thioredoxin |


|
 -0.33807 -0.33807
| |
| Rv3887c | conserved membrane protein |

|
 -0.3365 -0.3365
| |
| Rv3512 | PE-PGRS family protein |

|
 -0.33542 -0.33542
| |
| Rv2591 | PE-PGRS family protein |

|
 -0.33531 -0.33531
| |
| Rv1655 | acetylornithine aminotransferase argD |

|
 -0.33483 -0.33483
| |
| Rv2191 | conserved hypothetical protein |

|
 -0.33048 -0.33048
| |
| Rv1452c | PE-PGRS family protein |

|
 -0.32829 -0.32829
| |
| Rv2117 | conserved hypothetical protein |

|
 -0.32758 -0.32758
| |
| Rv1209 | conserved hypothetical protein |

|
 -0.32716 -0.32716
| |
| Rv2009 | conserved hypothetical protein |

|
 -0.32533 -0.32533
| |
| Rv0015c | transmembrane serine/threonine-protein kinase A pknA |

|
 -0.31866 -0.31866
| |
| Rv1215c | conserved hypothetical protein |

|
 -0.31821 -0.31821
| |
| Rv3737 | conserved membrane protein |

|
 -0.31817 -0.31817
| |
| Rv0668 | DNA-directed RNA polymerase beta chain rpoC |

|
 -0.31507 -0.31507
| |
| Rv2417c | conserved hypothetical protein |

|
 -0.31347 -0.31347
| |
| Rv3511 | PE-PGRS family protein |

|
 -0.31321 -0.31321
| |
| Rv2862c | conserved hypothetical protein |

|
 -0.3117 -0.3117
| |
| Rv2509 | short-chain type dehydrogenase/reductase |

|
 -0.30902 -0.30902
| |
| Rv0624 | conserved hypothetical protein |

|
 -0.30819 -0.30819
| |
| Rv1173 | F420 biosynthesis protein fbiC |


|
 -0.3074 -0.3074
| |
| Rv2345 | conserved membrane protein |

|
 -0.30209 -0.30209
| |
| Rv3508 | PE-PGRS family protein |

|
 -0.30097 -0.30097
| |
| Rv2439c | glutamate 5-kinase protein proB |

|
 -0.3 -0.3
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3634c | UDP-glucose 4-epimerase galE1 |


|
 0.48058 0.48058
| |
| Rv1575 | phiRv1 phage protein |

|
 0.45235 0.45235
| |
| Rv0620 | galactokinase galK |

|
 0.42662 0.42662
| |
| Rv2683 | conserved hypothetical protein |

|
 0.42474 0.42474
| |
| Rv1254 | acyltransferase |

|
 0.42202 0.42202
| |
| Rv0962c | lipoprotein lprP |

|
 0.42025 0.42025
| |
| Rv2262c | conserved hypothetical protein |

|
 0.4198 0.4198
| |
| Rv2240c | hypothetical protein |

|
 0.41896 0.41896
| |
| Rv2810c | transposase |

|
 0.41855 0.41855
| |
| Rv0518 | hypothetical exported protein |

|
 0.41776 0.41776
| |
| Rv0816c | thioredoxin thiX |


|
 0.40929 0.40929
| |
| Rv0277c | conserved hypothetical protein |

|
 0.40842 0.40842
| |
| Rv0970 | conserved membrane protein |

|
 0.40707 0.40707
| |
| Rv0344c | lipoprotein lpqJ |

|
 0.39172 0.39172
| |
| Rv3714c | conserved hypothetical protein |

|
 0.38872 0.38872
| |
| Rv0770 | dehydrogenase/reductase |

|
 0.38807 0.38807
| |
| Rv0052 | conserved hypothetical protein |

|
 0.38752 0.38752
| |
| Rv0130 | conserved hypothetical protein |

|
 0.38483 0.38483
| |
| Rv1582c | phiRv1 phage protein |

|
 0.38409 0.38409
| |
| Rv3488 | conserved hypothetical protein |

|
 0.38163 0.38163
| |
| Rv0224c | methyltransferase/methylase |


|
 0.37988 0.37988
| |
| Rv0982 | two component system sensor kinase mprB |

|
 0.3794 0.3794
| |
| Rv3490 | alpha, alpha-trehalose-phosphate synthase otsA |

|
 0.37192 0.37192
| |
| Rv2491 | conserved hypothetical protein |

|
 0.36995 0.36995
| |
| Rv0334 | alpha-D-glucose-1-phosphate thymidylyltransferase rmlA |

|
 0.36904 0.36904
| |
| Rv0977 | PE-PGRS family protein |

|
 0.36875 0.36875
| |
| Rv0810c | conserved hypothetical protein |

|
 0.36834 0.36834
| |
| Rv0966c | conserved hypothetical protein |

|
 0.36664 0.36664
| |
| Rv1888c | transmembrane protein |

|
 0.36563 0.36563
| |
| Rv0779c | conserved membrane protein |

|
 0.36435 0.36435
| |
| Rv2842c | conserved hypothetical protein |

|
 0.36355 0.36355
| |
| Rv2485c | carboxylesterase lipQ |

|
 0.36144 0.36144
| |
| Rv1972 | MCE-associated membrane protein |

|
 0.35847 0.35847
| |
| Rv2042c | conserved hypothetical protein |

|
 0.35726 0.35726
| |
| Rv1212c | glycosyltransferase |

|
 0.35609 0.35609
| |
| Rv0978c | PE-PGRS family protein |

|
 0.35608 0.35608
| |
| Rv2384 | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase mbtA |

|
 0.35494 0.35494
| |
| Rv2408 | PE family protein |

|
 0.35455 0.35455
| |
| Rv0991c | conserved serine rich protein |

|
 0.35185 0.35185
| |
| Rv0967 | conserved hypothetical protein |

|
 0.35068 0.35068
| |
| Rv1388 | integration host factor mihF |

|
 0.34965 0.34965
| |
| Rv2740 | conserved hypothetical protein |

|
 0.34938 0.34938
| |
| Rv1029 | potassium-transporting ATPase A chain kdpA |

|
 0.34871 0.34871
| |
| Rv0228 | membrane acyltransferase |

|
 0.34825 0.34825
| |
| Rv0318c | conserved membrane protein |

|
 0.34477 0.34477
| |
| Rv2090 | 5-3 exonuclease |

|
 0.34358 0.34358
| |
| Rv2416c | enhanced intracellular survival protein eis |


|
 0.33979 0.33979
| |
| Rv1031 | potassium-transporting ATPase C chain kdpC |

|
 0.33937 0.33937
| |
| Rv1004c | membrane protein |

|
 0.33783 0.33783
| |
| Rv0120c | elongation factor G fusA2 |

|
 0.33689 0.33689
| |
| Rv1670 | conserved hypothetical protein |

|
 0.33639 0.33639
| |
| Rv2742c | conserved arginine rich protein |

|
 0.33395 0.33395
| |
| Rv1672c | conserved membrane transport protein |




|
 0.33338 0.33338
| |
| Rv0680c | conserved membrane protein |

|
 0.33323 0.33323
| |
| Rv2679 | enoyl-CoA hydratase echA15 |

|
 0.33066 0.33066
| |
| Rv2872 | conserved hypothetical protein |

|
 0.33057 0.33057
| |
| Rv1954c | hypothetical protein |

|
 0.32837 0.32837
| |
| Rv0989c | polyprenyl-diphosphate synthase grcC2 |

|
 0.32655 0.32655
| |
| Rv1878 | glutamine synthetase glnA3 |

|
 0.32644 0.32644
| |
| Rv1290c | conserved hypothetical protein |

|
 0.32552 0.32552
| |
| Rv1546 | conserved hypothetical protein |

|
 0.32398 0.32398
| |
| Rv0504c | conserved hypothetical protein |

|
 0.32324 0.32324
| |
| Rv0891c | transcriptional regulator |

|
 0.32318 0.32318
| |
| Rv0368c | conserved hypothetical protein |

|
 0.32201 0.32201
| |
| Rv2040c | sugar-transport membrane protein ABC transporter |

|
 0.32158 0.32158
| |
| Rv1903 | conserved membrane protein |

|
 0.32025 0.32025
| |
| Rv0261c | membrane nitrite extrusion protein narK3 |

|
 0.31891 0.31891
| |
| Rv2094c | SEC-independent protein translocase membrane-bound protein tatA |

|
 0.31871 0.31871
| |
| Rv0339c | transcriptional regulator |

|
 0.31863 0.31863
| |
| Rv0749 | conserved hypothetical protein |

|
 0.3185 0.3185
| |
| Rv1752 | conserved hypothetical protein |

|
 0.31789 0.31789
| |
| Rv0380c | RNA methyltransferase |

|
 0.31705 0.31705
| |
| Rv1454c | quinone reductase qor |


|
 0.31628 0.31628
| |
| Rv0168 | hypothetical membrane protein yrbE1B |

|
 0.31614 0.31614
| |
| Rv1868 | conserved hypothetical protein |


|
 0.31334 0.31334
| |
| Rv2261c | conserved hypothetical protein |

|
 0.31278 0.31278
| |
| Rv2170 | conserved hypothetical protein |

|
 0.31277 0.31277
| |
| Rv1720c | conserved hypothetical protein |

|
 0.31261 0.31261
| |
| Rv0941c | conserved hypothetical protein |

|
 0.31065 0.31065
| |
| Rv2236c | cobalamin biosynthesis transmembrane protein cobD |

|
 0.31035 0.31035
| |
| Rv1722 | carboxylase |

|
 0.30941 0.30941
| |
| Rv1777 | cytochrome P450 144 cyp144 |

|
 0.30924 0.30924
| |
| Rv2093c | SEC-independent protein translocase transmembrane protein tatC |

|
 0.3089 0.3089
| |
| Rv2328 | PE family protein |

|
 0.30817 0.30817
| |
| Rv0150c | conserved hypothetical protein |

|
 0.30806 0.30806
| |
| Rv2612c | phosphatidylinositol synthase pgsA1 |

|
 0.30803 0.30803
| |
| Rv2285 | conserved hypothetical protein |

|
 0.30789 0.30789
| |
| Rv1469 | cation transporter P-type ATPase D ctpD |

|
 0.30738 0.30738
| |
| Rv1537 | DNA polymerase IV dinX |

|
 0.30687 0.30687
| |
| Rv3366 | tRNA/rRNA methylase spoU |

|
 0.30678 0.30678
| |
| Rv3637 | transposase |

|
 0.30632 0.30632
| |
| Rv3386 | transposase |

|
 0.30609 0.30609
| |
| Rv3789 | conserved membrane protein |

|
 0.30554 0.30554
| |
| Rv0456c | enoyl-CoA hydratase echA2 |

|
 0.30337 0.30337
| |
| Rv1979c | permease |

|
 0.30152 0.30152
| |
| Rv1674c | transcriptional regulator |

|
 0.30067 0.30067
| |
| Rv2566 | conserved hypothetical protein |

|
 0.30059 0.30059
|