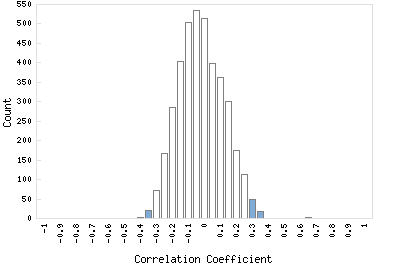
Correlation Catalog M. tuberculosis H37Rv: Rv1654 - acetylglutamate kinase argB
Distribution of gene correlation: Rv1654 vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2021c | transcriptional regulator |

|
 -0.38027 -0.38027
| |
| Rv2295 | conserved hypothetical protein |

|
 -0.372 -0.372
| |
| Rv2716 | conserved hypothetical protein |

|
 -0.35166 -0.35166
| |
| Rv2339 | transmembrane transport protein mmpL9 |

|
 -0.34845 -0.34845
| |
| Rv3847 | hypothetical protein |

|
 -0.33281 -0.33281
| |
| Rv2114 | hypothetical protein |

|
 -0.3309 -0.3309
| |
| Rv2633c | hypothetical protein |

|
 -0.32807 -0.32807
| |
| Rv2067c | conserved hypothetical protein |


|
 -0.32625 -0.32625
| |
| Rv1951c | conserved hypothetical protein |

|
 -0.32581 -0.32581
| |
| Rv1365c | anti-anti-sigma factor rsfA |

|
 -0.32245 -0.32245
| |
| Rv2075c | hypothetical exported protein |

|
 -0.31833 -0.31833
| |
| Rv1857 | molybdate-binding lipoprotein modA |

|
 -0.31803 -0.31803
| |
| Rv3849 | conserved hypothetical protein |

|
 -0.31668 -0.31668
| |
| Rv1952 | conserved hypothetical protein |

|
 -0.31457 -0.31457
| |
| Rv1041c | transposase |

|
 -0.31448 -0.31448
| |
| Rv2843 | conserved alanine rich membrane protein |

|
 -0.31006 -0.31006
| |
| Rv0741 | transposase |

|
 -0.30769 -0.30769
| |
| Rv0643c | methoxy mycolic acid synthase 3 mmaA3 |

|
 -0.30669 -0.30669
| |
| Rv1418 | lipoprotein lprH |

|
 -0.3054 -0.3054
| |
| Rv2949c | conserved hypothetical protein |

|
 -0.30418 -0.30418
| |
| Rv1535 | hypothetical protein |

|
 -0.30142 -0.30142
| |
| Rv2421c | nicotinate-nucleotide adenylyltransferase nadD |

|
 -0.30049 -0.30049
| |
| Rv3715c | recombination protein recR |

|
 -0.30002 -0.30002
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1652 | N-acetl-gamma-glutamyl-phoshate reductase argC |

|
 0.67313 0.67313
| |
| Rv1656 | ornithine carbamoyltransferase argF |

|
 0.66069 0.66069
| |
| Rv1653 | glutamate n-acetyltransferase argJ |

|
 0.58355 0.58355
| |
| Rv2614c | threonyl-tRNA synthetase thrS |

|
 0.45871 0.45871
| |
| Rv3306c | amidohydrolase amiB1 |

|
 0.42147 0.42147
| |
| Rv1916 | isocitrate lyase aceAb |

|
 0.39401 0.39401
| |
| Rv1126c | conserved hypothetical protein |

|
 0.39245 0.39245
| |
| Rv1480 | conserved hypothetical protein |

|
 0.3911 0.3911
| |
| Rv3626c | conserved hypothetical protein |

|
 0.38936 0.38936
| |
| Rv2448c | valyl-tRNA synthase protein valS |

|
 0.38211 0.38211
| |
| Rv2496c | pyruvate dehydrogenase E1 component beta subunit pdhB |

|
 0.3814 0.3814
| |
| Rv3800c | polyketide synthase pks13 |

|
 0.38009 0.38009
| |
| Rv1657 | arginine repressor argR |

|
 0.37845 0.37845
| |
| Rv2620c | conserved membrane protein |

|
 0.36829 0.36829
| |
| Rv0960 | conserved hypothetical protein |

|
 0.36201 0.36201
| |
| Rv0975c | acyl-CoA dehydrogenase fadE13 |

|
 0.36158 0.36158
| |
| Rv2610c | alpha-mannosyltransferase pimA |

|
 0.3591 0.3591
| |
| Rv1382 | hypothetical exported protein |

|
 0.35772 0.35772
| |
| Rv1474c | transcriptional regulator |

|
 0.35751 0.35751
| |
| Rv2110c | proteasome beta subunit prcB |

|
 0.3572 0.3572
| |
| Rv2992c | glutamyl-tRNA synthetase gltS |

|
 0.35428 0.35428
| |
| Rv1602 | amidotransferase hisH |

|
 0.35238 0.35238
| |
| Rv0568 | cytochrome P450 135B1 cyp135B1 |

|
 0.35045 0.35045
| |
| Rv1294 | homoserine dehydrogenase thrA |

|
 0.34813 0.34813
| |
| Rv1296 | homoserine kinase thrB |

|
 0.34785 0.34785
| |
| Rv0410c | serine/threonine-protein kinase pknG |

|
 0.34558 0.34558
| |
| Rv1655 | acetylornithine aminotransferase argD |

|
 0.34334 0.34334
| |
| Rv2678c | uroporphyrinogen decarboxylase hemE |

|
 0.3432 0.3432
| |
| Rv0411c | glutamine-binding lipoprotein glnH |

|
 0.34298 0.34298
| |
| Rv1462 | conserved hypothetical protein |

|
 0.34237 0.34237
| |
| Rv0484c | short-chain type oxidoreductase |

|
 0.34138 0.34138
| |
| Rv3596c | ATP-dependent protease ATP-binding subunit clpC1 |

|
 0.34071 0.34071
| |
| Rv1606 | phosphoribosyl-AMP-1,6-cyclohydrolase hisI |

|
 0.33887 0.33887
| |
| Rv2084 | hypothetical protein |

|
 0.3372 0.3372
| |
| Rv0752c | acyl-CoA dehydrogenase fadE9 |

|
 0.33687 0.33687
| |
| Rv3042c | phosphoserine phosphatase serB2 |

|
 0.33685 0.33685
| |
| Rv1086 | short-chain Z-isoprenyl diphosphate synthase |

|
 0.33684 0.33684
| |
| Rv1463 | ABC transporter ATP-binding protein |

|
 0.33616 0.33616
| |
| Rv2602 | conserved hypothetical protein |

|
 0.33355 0.33355
| |
| Rv1915 | isocitrate lyase aceAa |

|
 0.33255 0.33255
| |
| Rv3040c | conserved hypothetical protein |


|
 0.33065 0.33065
| |
| Rv1604 | inositol-monophosphatase impA |

|
 0.33061 0.33061
| |
| Rv2112c | conserved hypothetical protein |

|
 0.33049 0.33049
| |
| Rv2438c | glutamine-dependent NAD(+) synthetase nadE |

|
 0.33013 0.33013
| |
| Rv0382c | orotate phosphoribosyltransferase pyrE |

|
 0.32963 0.32963
| |
| Rv3814c | acyltransferase |

|
 0.32707 0.32707
| |
| Rv1122 | 6-phosphogluconate dehydrogenase, decarboxylating gnd2 |

|
 0.32637 0.32637
| |
| Rv1422 | conserved hypothetical protein |

|
 0.32368 0.32368
| |
| Rv3256c | conserved hypothetical protein |

|
 0.32356 0.32356
| |
| Rv0495c | conserved hypothetical protein |

|
 0.3209 0.3209
| |
| Rv0688 | ferredoxin reductase |

|
 0.32081 0.32081
| |
| Rv1465 | nitrogen fixation related protein |

|
 0.32039 0.32039
| |
| Rv2280 | dehydrogenase |

|
 0.31801 0.31801
| |
| Rv3014c | NAD-dependent DNA ligase ligA |

|
 0.31688 0.31688
| |
| Rv2794c | conserved hypothetical protein |

|
 0.31589 0.31589
| |
| Rv1466 | conserved hypothetical protein |

|
 0.31495 0.31495
| |
| Rv2495c | dihydrolipoamide S-acetyltransferase E2 component pdhC |

|
 0.3145 0.3145
| |
| Rv2454c | oxidoreductase beta subunit |

|
 0.31309 0.31309
| |
| Rv0038 | conserved hypothetical protein |

|
 0.31157 0.31157
| |
| Rv0356c | conserved hypothetical protein |

|
 0.31035 0.31035
| |
| Rv2064 | cobalamin biosynthesis protein cobG |

|
 0.31022 0.31022
| |
| Rv1072 | conserved membrane protein |

|
 0.30885 0.30885
| |
| Rv3264c | D-alpha-D-mannose-1-phosphate guanylyltransferase manB |


|
 0.30835 0.30835
| |
| Rv0724 | protease IV sppA |


|
 0.30826 0.30826
| |
| Rv3238c | conserved membrane protein |

|
 0.30769 0.30769
| |
| Rv1700 | conserved hypothetical protein |


|
 0.30698 0.30698
| |
| Rv3568c | biphenyl-2,3-diol 1,2-dioxygenase bphC |

|
 0.30548 0.30548
| |
| Rv0758 | two component system sensor kinase phoR |

|
 0.30419 0.30419
| |
| Rv0412c | conserved membrane protein |

|
 0.30411 0.30411
| |
| Rv2097c | conserved hypothetical protein |

|
 0.30301 0.30301
| |
| Rv3618 | monooxygenase |

|
 0.30178 0.30178
| |
| Rv3627c | conserved hypothetical protein |

|
 0.30156 0.30156
| |
| Rv3210c | conserved hypothetical protein |

|
 0.30001 0.30001
|