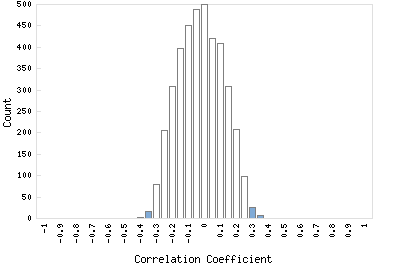
Correlation Catalog M. tuberculosis H37Rv: Rv1645c - conserved hypothetical protein
Distribution of gene correlation: Rv1645c vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2992c | glutamyl-tRNA synthetase gltS |

|
 -0.36747 -0.36747
| |
| Rv0058 | replicative DNA helicase dnaB |

|
 -0.36171 -0.36171
| |
| Rv1304 | ATP synthase A chain atpB |

|
 -0.3419 -0.3419
| |
| Rv3874 | 10 kda culture filtrate antigen esxB |

|
 -0.34115 -0.34115
| |
| Rv0734 | methionine aminopeptidase mapA |

|
 -0.3404 -0.3404
| |
| Rv2882c | ribosome recycling factor frr |

|
 -0.33893 -0.33893
| |
| Rv1307 | ATP synthase delta chain atpH |

|
 -0.33152 -0.33152
| |
| Rv1306 | ATP synthase B chain atpF |

|
 -0.32492 -0.32492
| |
| Rv0036c | conserved hypothetical protein |

|
 -0.31841 -0.31841
| |
| Rv0934 | periplasmic phosphate-binding lipoprotein pstS1 |

|
 -0.31228 -0.31228
| |
| Rv3648c | cold shock protein A cspA |

|
 -0.31103 -0.31103
| |
| Rv2228c | conserved hypothetical protein |

|
 -0.30931 -0.30931
| |
| Rv2082 | conserved hypothetical protein |

|
 -0.3073 -0.3073
| |
| Rv2790c | lipid-transfer protein ltp1 |

|
 -0.30672 -0.30672
| |
| Rv2604c | conserved hypothetical protein |

|
 -0.3056 -0.3056
| |
| Rv3718c | conserved hypothetical protein |

|
 -0.30414 -0.30414
| |
| Rv1078 | proline-rich antigen pra |

|
 -0.30309 -0.30309
| |
| Rv3198c | ATP-dependent DNA helicase II uvrD2 |

|
 -0.30176 -0.30176
| |
| Rv1070c | enoyl-CoA hydratase echA8 |

|
 -0.30059 -0.30059
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2661c | hypothetical protein |

|
 0.39476 0.39476
| |
| Rv1727 | conserved hypothetical protein |

|
 0.38621 0.38621
| |
| Rv0331 | dehydrogenase/reductase |

|
 0.37889 0.37889
| |
| Rv0335c | PE family protein |

|
 0.36201 0.36201
| |
| Rv0575c | oxidoreductase |


|
 0.35544 0.35544
| |
| Rv0826 | conserved hypothetical protein |

|
 0.35542 0.35542
| |
| Rv2643 | arsenic-transport membrane protein arsC |

|
 0.35329 0.35329
| |
| Rv0654 | dioxygenase |

|
 0.34495 0.34495
| |
| Rv3504 | acyl-CoA dehydrogenase fadE26 |

|
 0.34029 0.34029
| |
| Rv1669 | hypothetical protein |

|
 0.3396 0.3396
| |
| Rv2016 | hypothetical protein |

|
 0.33951 0.33951
| |
| Rv2642 | transcriptional regulator, arsR-family |

|
 0.3394 0.3394
| |
| Rv2641 | cadmium inducible protein cadI |

|
 0.33827 0.33827
| |
| Rv2665 | hypothetical arginine rich protein |

|
 0.33378 0.33378
| |
| Rv3054c | conserved hypothetical protein |

|
 0.33103 0.33103
| |
| Rv1673c | conserved hypothetical protein |

|
 0.32637 0.32637
| |
| Rv2877c | conserved membrane protein |

|
 0.31961 0.31961
| |
| Rv0573c | conserved hypothetical protein |

|
 0.31923 0.31923
| |
| Rv0753c | methylmalonate-semialdehyde dehydrogenase mmsA |

|
 0.31826 0.31826
| |
| Rv2797c | conserved hypothetical protein |

|
 0.31776 0.31776
| |
| Rv0656c | conserved hypothetical protein |

|
 0.31728 0.31728
| |
| Rv0267 | membrane nitrite extrusion protein narU |

|
 0.31664 0.31664
| |
| Rv0998 | conserved hypothetical protein |

|
 0.31478 0.31478
| |
| Rv1992c | metal cation transporting P-type ATPase ctpG |

|
 0.31371 0.31371
| |
| Rv1993c | conserved hypothetical protein |

|
 0.31081 0.31081
| |
| Rv3641c | cell filamentation protein fic |

|
 0.30963 0.30963
| |
| Rv1861 | conserved membrane protein |

|
 0.3095 0.3095
| |
| Rv3387 | transposase |

|
 0.30757 0.30757
| |
| Rv2639c | conserved membrane protein |

|
 0.30274 0.30274
| |
| Rv3617 | epoxide hydrolase ephA |

|
 0.30211 0.30211
| |
| Rv3124 | transcriptional regulator |

|
 0.30177 0.30177
| |
| Rv0767c | conserved hypothetical protein |

|
 0.30086 0.30086
|