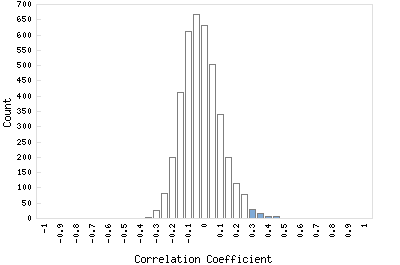
Correlation Catalog M. tuberculosis H37Rv: Rv0373c - carbon monoxyde dehydrogenase large chain
Distribution of gene correlation: Rv0373c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with positive correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0978c | PE-PGRS family protein |

|
 0.47637 0.47637
| |
| Rv0376c | conserved hypothetical protein |

|
 0.4713 0.4713
| |
| Rv0987 | adhesion component transport transmembrane protein ABC transporter |

|
 0.45777 0.45777
| |
| Rv0967 | conserved hypothetical protein |

|
 0.45584 0.45584
| |
| Rv0991c | conserved serine rich protein |

|
 0.45003 0.45003
| |
| Rv0982 | two component system sensor kinase mprB |

|
 0.44904 0.44904
| |
| Rv0326 | hypothetical protein |


|
 0.44844 0.44844
| |
| Rv0377 | transcriptional regulator, lysR-family |

|
 0.43553 0.43553
| |
| Rv0371c | conserved hypothetical protein |

|
 0.42482 0.42482
| |
| Rv0318c | conserved membrane protein |

|
 0.42347 0.42347
| |
| Rv1147 | conserved hypothetical protein |


|
 0.42088 0.42088
| |
| Rv0160c | PE family protein |

|
 0.40407 0.40407
| |
| Rv0979c | hypothetical protein |

|
 0.39383 0.39383
| |
| Rv0745 | conserved hypothetical protein |

|
 0.38818 0.38818
| |
| Rv1359 | transcriptional regulator |

|
 0.38804 0.38804
| |
| Rv0381c | hypothetical protein |

|
 0.38769 0.38769
| |
| Rv0093c | conserved membrane protein |

|
 0.38245 0.38245
| |
| Rv0600c | two component system sensor kinase |

|
 0.38228 0.38228
| |
| Rv2078 | hypothetical protein |

|
 0.38185 0.38185
| |
| Rv0362 | Mg2+ transport transmembrane protein mgtE |

|
 0.37789 0.37789
| |
| Rv0356c | conserved hypothetical protein |

|
 0.37579 0.37579
| |
| Rv0320 | hypothetical exported protein |

|
 0.36936 0.36936
| |
| Rv0152c | PE family protein |

|
 0.36763 0.36763
| |
| Rv0619 | galactose-1-phosphate uridylyltransferase galTb |

|
 0.36686 0.36686
| |
| Rv0735 | alternative RNA polymerase sigma factor sigL |

|
 0.35981 0.35981
| |
| Rv0825c | conserved hypothetical protein |

|
 0.35601 0.35601
| |
| Rv1120c | conserved hypothetical protein |

|
 0.35464 0.35464
| |
| Rv0980c | PE-PGRS family protein |

|
 0.35351 0.35351
| |
| Rv0416 | hypothetical protein thiS |

|
 0.34956 0.34956
| |
| Rv0368c | conserved hypothetical protein |

|
 0.3494 0.3494
| |
| Rv0393 | 13E12 repeat family protein |

|
 0.34857 0.34857
| |
| Rv0962c | lipoprotein lprP |

|
 0.34857 0.34857
| |
| Rv0691c | transcriptional regulator |

|
 0.34435 0.34435
| |
| Rv1228 | lipoprotein lpqX |

|
 0.34099 0.34099
| |
| Rv0456c | enoyl-CoA hydratase echA2 |

|
 0.33661 0.33661
| |
| Rv0699 | hypothetical protein |

|
 0.3353 0.3353
| |
| Rv1188 | proline dehydrogenase |

|
 0.3353 0.3353
| |
| Rv0209 | hypothetical protein |

|
 0.33418 0.33418
| |
| Rv2423 | hypothetical protein |


|
 0.3335 0.3335
| |
| Rv1145 | transmembrane transport protein mmpL13a |

|
 0.33163 0.33163
| |
| Rv3439c | conserved alanine and proline rich protein |

|
 0.33097 0.33097
| |
| Rv0755c | PPE family protein |

|
 0.32704 0.32704
| |
| Rv0218 | conserved membrane protein |

|
 0.32068 0.32068
| |
| Rv0459 | conserved hypothetical protein |

|
 0.31946 0.31946
| |
| Rv0193c | hypothetical protein |

|
 0.31769 0.31769
| |
| Rv0768 | aldehyde dehydrogenase NAD-dependent aldA |

|
 0.3156 0.3156
| |
| Rv0294 | trans-aconitate methyltransferase tam |


|
 0.31533 0.31533
| |
| Rv0215c | acyl-CoA dehydrogenase fadE3 |

|
 0.31364 0.31364
| |
| Rv0347 | conserved membrane protein |

|
 0.31176 0.31176
| |
| Rv0964c | hypothetical protein |

|
 0.31 0.31
| |
| Rv0121c | conserved hypothetical protein |

|
 0.30686 0.30686
| |
| Rv0098 | conserved hypothetical protein |

|
 0.30557 0.30557
| |
| Rv0252 | nitrite reductase large subunit nirB |

|
 0.3042 0.3042
| |
| Rv0965c | conserved hypothetical protein |

|
 0.30371 0.30371
| |
| Rv0096 | PPE family protein |

|
 0.30302 0.30302
| |
| Rv0597c | conserved hypothetical protein |

|
 0.30206 0.30206
| |
| Rv0382c | orotate phosphoribosyltransferase pyrE |

|
 0.30173 0.30173
|