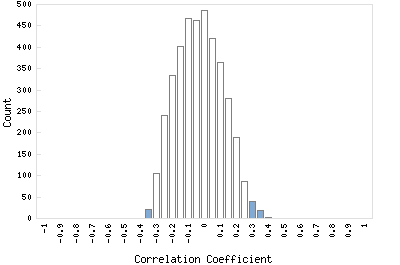
Correlation Catalog M. tuberculosis H37Rv: Rv0254c - bifunctional cobalamin biosynthesis protein cobU
Distribution of gene correlation: Rv0254c vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2700 | conserved secreted protein |

|
 -0.32582 -0.32582
| |
| Rv3804c | secreted fibronectin-binding protein antigen fbpA |

|
 -0.32214 -0.32214
| |
| Rv3106 | NADPH:adrenodoxin oxidoreductase fprA |


|
 -0.32125 -0.32125
| |
| Rv2477c | macrolide-transport ATP-binding protein ABC transporter |

|
 -0.31939 -0.31939
| |
| Rv2244 | meromycolate extension acyl carrier protein acpM |


|
 -0.31506 -0.31506
| |
| Rv3116 | molybdenum cofactor biosynthesis protein moeB2 |

|
 -0.31312 -0.31312
| |
| Rv1595 | L-aspartate oxidase nadB |

|
 -0.3126 -0.3126
| |
| Rv3219 | transcriptional regulator whib-like whiB1 |

|
 -0.31024 -0.31024
| |
| Rv3028c | electron transfer flavoprotein alpha subunit fixB |

|
 -0.30822 -0.30822
| |
| Rv0706 | 50S ribosomal protein L22 rplV |

|
 -0.30745 -0.30745
| |
| Rv3220c | two component system sensor kinase |

|
 -0.30647 -0.30647
| |
| Rv0722 | 50S ribosomal protein L30 rpmD |

|
 -0.30564 -0.30564
| |
| Rv3030 | conserved hypothetical protein |


|
 -0.30308 -0.30308
| |
| Rv3285 | bifunctional acetyl-/propionyl-coenzyme A carboxylase alpha chain accA3 |

|
 -0.30304 -0.30304
| |
| Rv3265c | dTDP-rha:a-D-glcnac-diphosphoryl polyprenol, a-3-L-rhamnosyl transferase wbbL1 |

|
 -0.3023 -0.3023
| |
| Rv3396c | GMP synthase guaA |

|
 -0.30225 -0.30225
| |
| Rv3648c | cold shock protein A cspA |

|
 -0.30084 -0.30084
| |
| Rv3250c | rubredoxin rubB |

|
 -0.30037 -0.30037
| |
| Rv2785c | 30S ribosomal protein S15 rpsO |

|
 -0.30028 -0.30028
| |
| Rv2883c | uridylate kinase pyrH |

|
 -0.30007 -0.30007
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0614 | conserved hypothetical protein |

|
 0.51668 0.51668
| |
| Rv0115 | D-alpha-D-heptose-7-phosphate kinase hddA |

|
 0.46206 0.46206
| |
| Rv0521c |

|
 0.4406 0.4406
| ||
| Rv0376c | conserved hypothetical protein |

|
 0.43623 0.43623
| |
| Rv1000 |

|
 0.42264 0.42264
| ||
| Rv1089 | PE family protein |

|
 0.39815 0.39815
| |
| Rv0236c | conserved membrane protein |

|
 0.38536 0.38536
| |
| Rv1012 | hypothetical protein |

|
 0.38006 0.38006
| |
| Rv0106 | conserved hypothetical protein |

|
 0.37996 0.37996
| |
| Rv0096 | PPE family protein |

|
 0.37881 0.37881
| |
| Rv0600c | two component system sensor kinase |

|
 0.37694 0.37694
| |
| Rv0086 | hydrogenase hycQ |


|
 0.37083 0.37083
| |
| Rv0974c | acetyl-/propionyl-CoA carboxylase beta subunit accD2 |

|
 0.36942 0.36942
| |
| Rv0228 | membrane acyltransferase |

|
 0.36796 0.36796
| |
| Rv0224c | methyltransferase/methylase |


|
 0.36765 0.36765
| |
| Rv0278c | PE-PGRS family protein |

|
 0.36393 0.36393
| |
| Rv0162c | zinc-type alcohol dehydrogenase E subunit adhE |

|
 0.36364 0.36364
| |
| Rv1358 | transcriptional regulator |

|
 0.36231 0.36231
| |
| Rv1145 | transmembrane transport protein mmpL13a |

|
 0.36211 0.36211
| |
| Rv0258c | conserved hypothetical protein |

|
 0.36119 0.36119
| |
| Rv0042c | transcriptional regulator, marR-family |

|
 0.35641 0.35641
| |
| Rv0114 | D-alpha,beta-D-heptose-1,7-biphosphate phosphatase gmhB |

|
 0.35625 0.35625
| |
| Rv0449c | conserved hypothetical protein |

|
 0.35525 0.35525
| |
| Rv0570 | ribonucleoside-diphosphate reductase large subunit nrdZ |

|
 0.34998 0.34998
| |
| Rv0594 | MCE-family protein mce2F |

|
 0.34991 0.34991
| |
| Rv3109 | molybdenum cofactor biosynthesis protein A moaA1 |

|
 0.34973 0.34973
| |
| Rv0976c | conserved hypothetical protein |

|
 0.34843 0.34843
| |
| Rv0213c | methyltransferase/methylase |

|
 0.34768 0.34768
| |
| Rv0618 | galactose-1-phosphate uridylyltransferase galTa |

|
 0.34642 0.34642
| |
| Rv0765c | oxidoreductase |


|
 0.34539 0.34539
| |
| Rv0218 | conserved membrane protein |

|
 0.34395 0.34395
| |
| Rv0329c | conserved hypothetical protein |


|
 0.34337 0.34337
| |
| Rv0979c | hypothetical protein |

|
 0.34163 0.34163
| |
| Rv0619 | galactose-1-phosphate uridylyltransferase galTb |

|
 0.33984 0.33984
| |
| Rv0095c | conserved hypothetical protein |

|
 0.33775 0.33775
| |
| Rv0325 | hypothetical protein |

|
 0.33658 0.33658
| |
| Rv0209 | hypothetical protein |

|
 0.33475 0.33475
| |
| Rv0093c | conserved membrane protein |

|
 0.33421 0.33421
| |
| Rv1743 | transmembrane serine/threonine-protein kinase E pknE |

|
 0.32883 0.32883
| |
| Rv0087 | formate hydrogenase hycE |

|
 0.32531 0.32531
| |
| Rv0120c | elongation factor G fusA2 |

|
 0.32065 0.32065
| |
| Rv0109 | PE-PGRS family protein |

|
 0.32045 0.32045
| |
| Rv0459 | conserved hypothetical protein |

|
 0.31922 0.31922
| |
| Rv0121c | conserved hypothetical protein |

|
 0.31809 0.31809
| |
| Rv0620 | galactokinase galK |

|
 0.3179 0.3179
| |
| Rv0152c | PE family protein |

|
 0.31746 0.31746
| |
| Rv1530 | alcohol dehydrogenase adh |


|
 0.31727 0.31727
| |
| Rv2423 | hypothetical protein |


|
 0.31716 0.31716
| |
| Rv0253 | nitrite reductase small subunit nirD |


|
 0.31514 0.31514
| |
| Rv0362 | Mg2+ transport transmembrane protein mgtE |

|
 0.31476 0.31476
| |
| Rv0398c | secreted protein |

|
 0.3139 0.3139
| |
| Rv0508 | conserved hypothetical protein |


|
 0.31269 0.31269
| |
| Rv0494 | transcriptional regulator, gntR-family |

|
 0.31112 0.31112
| |
| Rv0252 | nitrite reductase large subunit nirB |

|
 0.31028 0.31028
| |
| Rv0573c | conserved hypothetical protein |

|
 0.30888 0.30888
| |
| Rv1052 | hypothetical protein |

|
 0.30861 0.30861
| |
| Rv0377 | transcriptional regulator, lysR-family |

|
 0.30717 0.30717
| |
| Rv0518 | hypothetical exported protein |

|
 0.3067 0.3067
| |
| Rv0603 | hypothetical exported protein |

|
 0.30655 0.30655
| |
| Rv2270 | lipoprotein lppN |

|
 0.30649 0.30649
| |
| Rv0882 | transmembrane protein |

|
 0.30594 0.30594
| |
| Rv0215c | acyl-CoA dehydrogenase fadE3 |

|
 0.30369 0.30369
| |
| Rv0262c | aminoglycoside 2-N-acetyltransferase aac |

|
 0.30202 0.30202
|